Ngā Rākau Taketake teams showcase their results
Conference season has returned at last! Despite the added facial attire, our plant pathogen teams made the most of being able to connect kanohi ki te kanohi at the recent New Zealand Plant Protection Society conference.
The event kicked off in Ōtautahi (Christchurch) with a special symposium: Plant pathogens that keep us awake: past, present and future threats to native species. Many Ngā Rākau Taketake researchers took the opportunity to present talks and posters giving updates on their research – here are a few highlights from the day.

Understanding the enemy: how does Austropuccinia psidii infect its host to cause myrtle rust?
Grant Smith (Plant & Food Research) reminded the audience that myrtle rust isn’t waiting around for us to learn about it.

“While we’re describing resistance [to the pathogen] at the plant level, the pathogen is still evolving. ‘Race 5’ is more aggressive than the first four races described from Brazil,” he said.
“And it has now been confirmed it is reproducing sexually.” This means it will be able to adapt to new environments easier than if it was cloning itself.
Grant and his team have researched the susceptibility of our native plant species in glasshouses (a controlled environment) – and the results are concerning.
“In terms of our taonga, when we put them through an intensive susceptibility test, we see no resistance in ramarama and rōhutu, and very little in pōhutukawa,” he said.
Some good news is that Grant and his team have found A. psidii shows the same gene expression (acts the same way) regardless of the resistance of the mānuka plant it lands on. This means that if we can stop or interrupt gene expression early in the infection process, the ‘programming’ of gene expression to infect the plant will be interrupted and we could stop the disease developing.
Grant said resistance testing of native plant species is much harder than in the more common horticultural setting because of the genetic diversity. Plants bred for horticulture are clones, which means they will all show the exact same response (susceptibility or resistance) to a pathogen infection.
The plants used in these experiments are from known mother plants, but they were naturally pollinated so researchers don’t know where the paternal genetics came from. The plants could either be half siblings (sharing 50% of their genes) or full siblings (sharing 100% of their genes). This means each individual within a plant family group can respond differently to infection. This is a huge benefit for the survival of the wild population, but makes genetic research a lot more difficult!
Towards management of myrtle rust in urban and forest environments

The lab and field are two very different places when it comes to fungal infection, so it’s essential to test susceptibility in both.
Rob Beresford (Plant & Food Research) said to date, most of the susceptibility information we have for myrtle rust has come from field observations, but researchers need to control as many variables as they can to ensure the observations they’re making aren’t caused by unknown factors.
To address this, in 2019 Rob and his collegues at Plant & Food Research and Scion planted field trials in Rotorua and Tāmaki Makaurau. Their aim was to test Myrtaceae susceptibility in a ‘controlled’ field environment.
With monthly monitoring over two years the team obseved no myrtle rust infection on kānuka and only one isolated incidence on mānuka fruit, but Rob said there was no dieback on either species.
Rōhutu were very slow to establish, and little growth meant little rust infection, but when the plants took off in the second year of the study their new growth became heavily infected.
The more favourable Auckland climate and soil resulted in higher growth rates in that study plot, which meant higher rates of myrtle rust infection.
Rob said “There were two distinct epidemics: on the spring growth flush and on the autumn re-growth.”
The team also made use of a cherry picker to examine natural infection of mature maire tawake. From the ground the canopies appeared intact, but Rob said this was a very different picture from above.
“Sadly all the young shoots and flowers on the plants were being killed [by myrtle rust]. That seemed to be the pattern in the tree we were closest to, and the others next to it.” Rob outlined a number of possible, place-specific controls, but said “The best hope to save threatened species in natural forest environments is genetic resistance. Resistant plants could be identified by mass planting of seedlings in field plots and selecting those that are less impacted by myrtle rust.”
The Phytophthora agathidicida genome sequence: a long-lasting resource for the research community
Earlier this year Rosie Bradshaw (Massey University) was part of a team who built what they think is the first chromosome-level Phytophthora genome assembly in the world.
The genome belongs to Phytophthora agathidicida and should help us to learn more about the pathogen that causes kauri dieback.

Rosie said “We’ve gone from a genome that is very fragmented, with lots of gaps, to one that is fully assembled to chromosome level and should contain all the genes.
“What we want to do now is find out which genes, and which molecules encoded by those genes, are important.”
‘Effectors’ are molecules secreted from pathogens that help to infect the host plant, but some of them are recognised by the plant immune system and trigger a defence response. If Rosie and her team can identify and characterise the most important P. agathidicida effectors, they could be used to screen for plant resistance or for the development of other kinds of disease management tools.
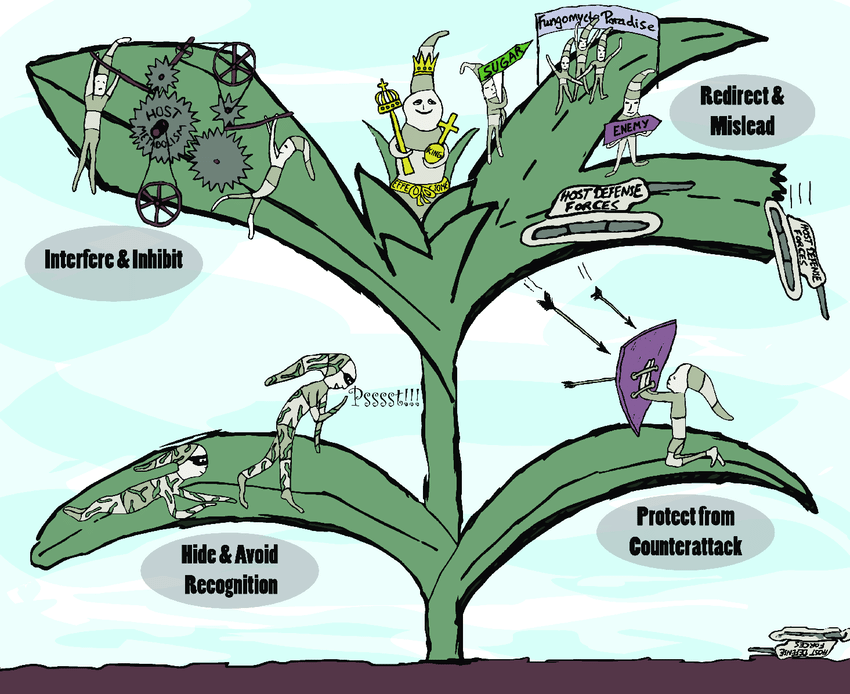
From their initial investigations researchers have found one particular effector of interest called RXLR29. This was not only present in the P. agathidicida genome but it was repeated nine times. It is able to trigger cell death, which means when the plant detects the effector, it will kill off cells around the infection to stop the pathogen travelling and infecting the entire plant.
This looked like good news, but Rosie said “There was zero expression.”
All the genes for the effector were either silenced or switched off, so the potential is there but it is never actually used. This has most likely occured to avoid detection by the host plant and help P. agathidicida to spread, and it highlights how adaptable this pathogen is.
This newly-detailed genome will be a resource for researchers for years to come – helping us learn more about P. agathidicida and how we might be able to control the pathogen at a genetic level.
Improving the understanding of kauri decline

Nari Williams (Plant & Food Research) said “As pathologists, we always come back to the disease triangle of ‘pathogen, host susceptibility and environment’. When we have all three we get disease, and when we get two we don’t necessarily get disease but we will certainly get confusion.”
If you have the pathogen and a susceptible host but no disease, what about the environment is preventing infection? If you have the right environment and a host, why is the disease not there?
With part of their research the Host, Pathogen & Environment team wanted to monitor the spread and intensification of kauri dieback over time. This would hopefully show patterns of infection and give some answers about how and why the pathogen was spreading.
They lined up aerial photos from as far back as 1942 to look at the state of the mature kauri, and have gotten a fair amount of detail considering the age of the photos.

Once the team saw patterns emerging in crown declines overtime, they started modelling them along with landscape factors, to better understand the rate of decline and disease impacts over time.
Nari said “The modelling is getting down to a 50m scale, which is almost useful for management . . . It’s a work in progress.
“Our preliminary results so far show canopy decline is associated with roads, tracks and waterways.”
The Te Roroa team are now doing tree and forest health monitoring surveys to validate these observations in the field and a similar approach is being implemented with Patuharakeke and Ngāti Rua.
Cultural monitoring: responding to kauri dieback and myrtle rust
A marine scientist by day, Kiri Reihana (Ngāpuhi, Te Rarawa, Tūhoe, Whakatōhea; Taiao Ora Solutions) has been moonlighting with the Control, Protect, Cure research theme and Ngāti Rangi to develop cultural indicators and metrics for kauri dieback and myrtle rust.
Working from an original app she designed called NGAHEREora (intended to monitor forest health), Kiri collaborated with mana whenua to extend the use of the tool to look at the effects of these two plant pathogens.

She said “It was a very Te Ao Māori approrach: looking at the health of the environment in its totality.”
Ngāti Rangi originally used indicators divided into five categories: Tangata, Wai, Rongoā, Manu and Ngahere. To make the tool specific for Ngā Rākau Taketake they added five more:
- Mauiuitanga – judging the sickness of the trees or any observations resulting from the sickness.
- The mauri of the environment – the water held within natural bodies such as the soil and the plants. The wetness of an environment is a good indicator of the mauri of that environment. For this tool the addition of the waterways was important indicator for ecosystem health.
- Rongo – what is the smell of the wai in the ngahere? Every field worker knows that the forest smells different at different times of the year and after events such as heavy rainfall. This can be used to indicate the state of the environment.
- Senses – are the senses of touch, smell, sound, sight and taste activated by the site?
- Can whānau exercise oranga? Are there enough kai or resources to support hui, such as tangihanga and the whānau?
Instead of an app (as with the original metrics), Kiri formatted these indicators into the suite of surveys for kauri dieback, myrtle rust, pathogen risk assessment and ecosystem impacts. These were created by the various research streams within Ngā Rākau Taketake. These surveys now form part of the suite of tools the research teams have developed to monitor kauri dieback and myrtle rust.
Why biosecurity needs the reflexive scientist
Since there are so many people involved in the biosecurity system, we need to find what holds us together, said social scientist Marie McEntee (University of Auckland).
“Relationship building is critical for effectively working within the complexity of biosecurity.

“Those of us who work in the biosecurity system need to reflect on who sits around the table. Collaboration requires dialogue to humanise relationships . . . it does not walk away from tension, it embraces it.”
The co-lead of Mobilising for Action (MfA) took the opportunity of her speaking slot to show how well widespread collaboration can work. The MfA research team operates within a waka hourua model, which enables mātauranga Māori and Western science to work alongside each other in understanding the human dimensions of forest health. Their transdiciplinary team includes social scientists (including many early career people), biological scientists, mana whenua, schools, communities, government agencies, NGOs and artists. Because of this they have made huge strides in their research and have just had an impressive 12 papers accepted for a special edition of the international journal Knowledge Cultures, due to be published early next year.
Marie said it is so important to be reflexive and transdisciplinary in our research.
Reflexivity means we question the assumptions and beliefs that drive how we operate as researchers. Transdisciplinarity iteratively weaves knowledge systems, skills, methodologies, values and fields of expertise. Marie said in Aotearoa this must be informed by Te Tiriti o Waitangi, kaupapa Māori, tikanga and mātauranga Māori.
“It is easy to fall into the culture of technical control, that limits who sits around the table,” she said.
“Everyone in this room is an asset to the biosecurity system. But perhaps the greatest threat to biosecurity also lies within us.”

Posted August 2022.